
- Bioactive Compounds
- By Signaling Pathways
- PI3K/Akt/mTOR
- Epigenetics
- Methylation
- Immunology & Inflammation
- Protein Tyrosine Kinase
- Angiogenesis
- Apoptosis
- Autophagy
- ER stress & UPR
- JAK/STAT
- MAPK
- Cytoskeletal Signaling
- Cell Cycle
- TGF-beta/Smad
- Compound Libraries
- Popular Compound Libraries
- Customize Library
- Clinical and FDA-approved Related
- Bioactive Compound Libraries
- Inhibitor Related
- Natural Product Related
- Metabolism Related
- Cell Death Related
- By Signaling Pathway
- By Disease
- Anti-infection and Antiviral Related
- Neuronal and Immunology Related
- Fragment and Covalent Related
- FDA-approved Drug Library
- FDA-approved & Passed Phase I Drug Library
- Preclinical/Clinical Compound Library
- Bioactive Compound Library-I
- Bioactive Compound Library-Ⅱ
- Kinase Inhibitor Library
- Express-Pick Library
- Natural Product Library
- Human Endogenous Metabolite Compound Library
- Alkaloid Compound LibraryNew
- Angiogenesis Related compound Library
- Anti-Aging Compound Library
- Anti-alzheimer Disease Compound Library
- Antibiotics compound Library
- Anti-cancer Compound Library
- Anti-cancer Compound Library-Ⅱ
- Anti-cancer Metabolism Compound Library
- Anti-Cardiovascular Disease Compound Library
- Anti-diabetic Compound Library
- Anti-infection Compound Library
- Antioxidant Compound Library
- Anti-parasitic Compound Library
- Antiviral Compound Library
- Apoptosis Compound Library
- Autophagy Compound Library
- Calcium Channel Blocker LibraryNew
- Cambridge Cancer Compound Library
- Carbohydrate Metabolism Compound LibraryNew
- Cell Cycle compound library
- CNS-Penetrant Compound Library
- Covalent Inhibitor Library
- Cytokine Inhibitor LibraryNew
- Cytoskeletal Signaling Pathway Compound Library
- DNA Damage/DNA Repair compound Library
- Drug-like Compound Library
- Endoplasmic Reticulum Stress Compound Library
- Epigenetics Compound Library
- Exosome Secretion Related Compound LibraryNew
- FDA-approved Anticancer Drug LibraryNew
- Ferroptosis Compound Library
- Flavonoid Compound Library
- Fragment Library
- Glutamine Metabolism Compound Library
- Glycolysis Compound Library
- GPCR Compound Library
- Gut Microbial Metabolite Library
- HIF-1 Signaling Pathway Compound Library
- Highly Selective Inhibitor Library
- Histone modification compound library
- HTS Library for Drug Discovery
- Human Hormone Related Compound LibraryNew
- Human Transcription Factor Compound LibraryNew
- Immunology/Inflammation Compound Library
- Inhibitor Library
- Ion Channel Ligand Library
- JAK/STAT compound library
- Lipid Metabolism Compound LibraryNew
- Macrocyclic Compound Library
- MAPK Inhibitor Library
- Medicine Food Homology Compound Library
- Metabolism Compound Library
- Methylation Compound Library
- Mouse Metabolite Compound LibraryNew
- Natural Organic Compound Library
- Neuronal Signaling Compound Library
- NF-κB Signaling Compound Library
- Nucleoside Analogue Library
- Obesity Compound Library
- Oxidative Stress Compound LibraryNew
- Plant Extract Library
- Phenotypic Screening Library
- PI3K/Akt Inhibitor Library
- Protease Inhibitor Library
- Protein-protein Interaction Inhibitor Library
- Pyroptosis Compound Library
- Small Molecule Immuno-Oncology Compound Library
- Mitochondria-Targeted Compound LibraryNew
- Stem Cell Differentiation Compound LibraryNew
- Stem Cell Signaling Compound Library
- Natural Phenol Compound LibraryNew
- Natural Terpenoid Compound LibraryNew
- TGF-beta/Smad compound library
- Traditional Chinese Medicine Library
- Tyrosine Kinase Inhibitor Library
- Ubiquitination Compound Library
-
Cherry Picking
You can personalize your library with chemicals from within Selleck's inventory. Build the right library for your research endeavors by choosing from compounds in all of our available libraries.
Please contact us at [email protected] to customize your library.
You could select:
- Antibodies
- Bioreagents
- qPCR
- 2x SYBR Green qPCR Master Mix
- 2x SYBR Green qPCR Master Mix(Low ROX)
- 2x SYBR Green qPCR Master Mix(High ROX)
- Protein Assay
- Protein A/G Magnetic Beads for IP
- Anti-Flag magnetic beads
- Anti-Flag Affinity Gel
- Anti-Myc magnetic beads
- Anti-HA magnetic beads
- Poly FLAG Peptide lyophilized powder
- Protease Inhibitor Cocktail
- Protease Inhibitor Cocktail (EDTA-Free, 100X in DMSO)
- Phosphatase Inhibitor Cocktail (2 Tubes, 100X)
- Cell Biology
- Cell Counting Kit-8 (CCK-8)
- Animal Experiment
- Mouse Direct PCR Kit (For Genotyping)
- New Products
- Contact Us
Mouse Direct PCR Kit (For Genotyping)
For research use only.
The Mouse Direct PCR Kit provides a fast preparation and PCRamplification that is specifically designed for mouse genotyping.

Selleck's has been cited by 63 publications
Price Comparison
Description
The Mouse Direct PCR Kit provides a fast preparation and PCRamplification that is specifically designed for mouse genotyping. TheBuffer L and Protease Plus rapidly digest mouse tail, ear and toe to release intact genomic DNA that can be used directly as the templatefor PCR amplification.By using this kit, the digestion process only takes 15 minutes. In addition, the 2x M-PCR OPTI Mix(which includes an optimized Taq Polymerase) ensures highamplification efficiency of target DNA.
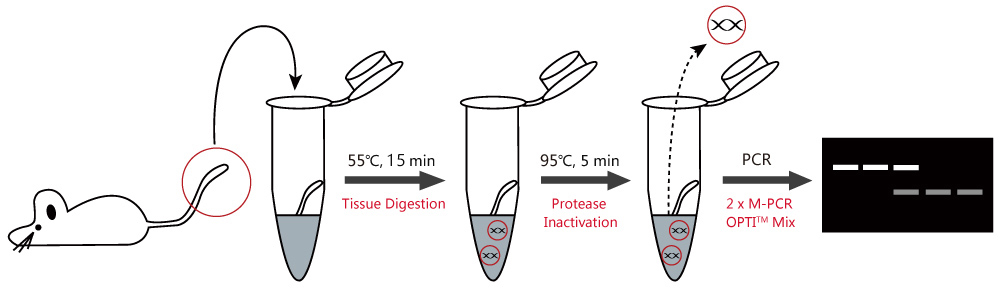
1. Comparison Between Different Methods
| Reagent | Price ($) | Genotyping Method | ||
|---|---|---|---|---|
| NaOH-HCL | Proteinase K | Selleck Direct PCR Kit | ||
| Lysis Buffer | Sigma, 0.010 | + | + | Buffer L |
| Proteinase K | Sigma P6556, 1U/reaction, 0.034 | + | Protease Plus | |
| Phenol Chloroform | Sigma P3803, 200μL/reaction, 0.116 | + | + | |
| Alcohols | Sigma I9030, 459836, 0.084 | + | + | |
| Taq | NEB M0273X, 1U/reaction, 0.122 | + | + | 2 x M-PCR OPTITM Mix |
| dNTP | Sigma GE28-4065-64, 0.25mM, 0.066 | + | + | |
| MgCL2, DNA Loading | Sigma, 0.010 | + | + | |
| Price ($) /Reaction | 0.41 | 0.44 | 0.39 | |
2. Comparison of PCR results (wild-type mouse)
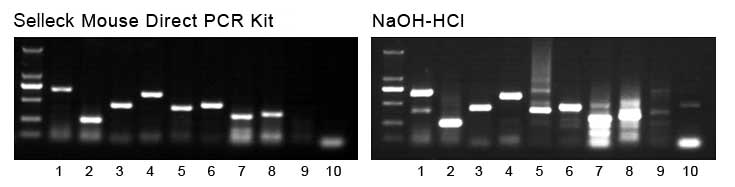
Lane 1-10 represents different PCR primers. Two of them serve as the negative controls (Lane 9 and 10) which only display bands when using the transgenic mouse sample. Therefore, Mouse Direct PCR kit ensures a higher PCR specificity, making it a reliable product for scientific research.
3. Advantages
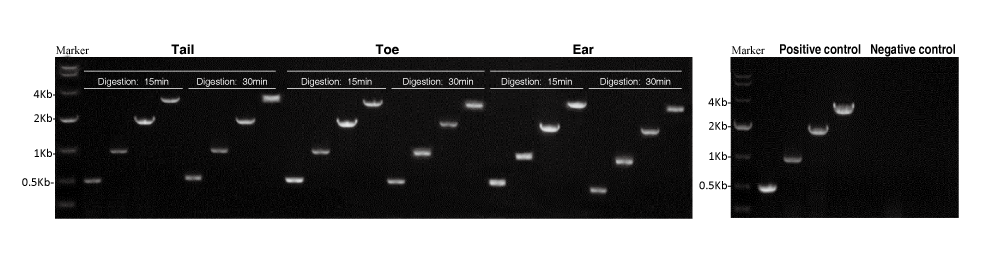
Analysis of Result:
The Mouse Direct PCR Kit applies to the preparation for samples from mouse tail, ear and toe. No significant difference was observed between the groups, which, digests in 55℃ for 15 min or 30 min. The kit is also effective for the amplification of large fragments (e.g. 3Kb).
Note: The primers for amplification of 500bp and 3Kb-fragments is designed based on the mouse GAPDH gene, while the primers for 1Kb and 2Kb-fragments is designed based on the mouse Ppip5k2 gene.
4. Components
| Complete Kit | B40013 (200 rxns) | B40015 (500 rxns) |
|---|---|---|
| Buffer L | 20 mL | 25 mL x 2 |
| Protease Plus | 0.4 mL | 1 mL |
| 2 x M-PCR OPTITM Mix (Dye Plus) | 1 mL x 2 | 1 mL x 5 |
| User Guide | 1 | 1 |
Buffer L: Lysis buffer.
Protease Plus: For rapid and efficient digestion of mouse tissue in only 15 minutes!
2x M-PCR OPTITM Mix: Includes Selleck's trademarked and optimized Taq DNA polymerase, dNTPs, MgCl2, and reaction buffer.
Storage (From the date of receipt)
2 x M-PCR OPTITM Mix and Protease Plus should be stored at -20°C for 2 years. If the PCR Mix is to be used frequently, it can be stored at 4°C for up to 10 days.
Buffer L should be stored at 4°C.
Protocol
1. Experimental Protocol
1) Genomic DNA Preparation
Place the mouse tail, ear, or toe in a 1.5 mL centrifuge tube. Thoroughly mix 100 μL of fresh Buffer L with 2 μL of Protease Plus for a single sample in a separate tube. Add the protease mixture to the mouse tissue tubes with the tissue cut submerged in it, then incubate at 55°C for 15 minutes. After the digestion process, incubate at 95°C for 5 minutes to inactivate protease. The tissue lysate can now be used as a PCR template.
2) PCR Genotyping
Add dd H2O, primers, template, and 2 x M-PCR OPTITM Mix into a PCR tube according to the recommended concentrations. Give the mixture a quick spin in the centrifuge and load into PCR amplifier to begin amplification. It is recommended to prepare PCR reaction in ice-bath.
| PCR Reaction Components | 20 μL Reaction Volume (μL) | 50 μL Reaction Volume (μL) |
|---|---|---|
| ddH2O | 8 | 21 |
| Forward Primer (10 μM) | 0.5 | 1 |
| Reverse Primer (10 μM) | 0.5 | 1 |
| Template | 1 | 2 |
| 2 x M-PCR OPTI™ Mix | 10 | 25 |
| PCR Steps | Temperature (°C) | Time | Cycles |
|---|---|---|---|
| 1 | 94 | 5 min | 1 |
| 2 | 94 | 20 sec | 35 |
| 3 | 50-65 | 30 sec | |
| 4 | 72 | X min (2 kb /min) | |
| 5 | 72 | 5 min | 1 |
| 6 | 12 | -- | 1 |
Note: Before First Use
1. All reagents in this kit have been optimized for use together and any modifications or alternative uses are prohibited.
2. For each step, make sure every reagent in the kit is blended well prior to use.
3. During the tissue digestion step, shaking the tubes 1-2 times will be helpful to release the genomic DNA.
4. For most mouse tissue samples, an incubation of 15 minutes at 55°C will suffice for genomic DNA extraction. The tissue may still appear intact, but extraction has occurred.
5. The extracted mouse genomic DNA should be applied immediately prior to the PCR amplification step. Inappropriate long-term storage may cause unreliable PCR amplifications.
Trouble Shooting
Please review the following for trouble-shooting options when you encounter technical difficulties. Alternatively, feel free to contact Selleck technical support directly.
| Problem | Potential Cause (s) | Suggestion (s) |
|---|---|---|
| No amplification product in test or control samples | Amplification reaction was incorrectly set up | Optimize the proper reaction set up |
| Improper storage has led to loss of activity of PCR reagents | Replace all components with fresh reagents | |
| Primers are not optimal and did not anneal | Redesign primers | |
| Amplification worked in the control samples, but not in test samples | Digestion was incomplete | Extend digestion time up to 30 minutes at 55°C |
| Samples were stored too long and genomic DNA degraded | Collect fresh mouse tail samples for genomic DNA extraction | |
| The lysis solution was mixed with PCR mixture for too long | PCR reactions should be initiated within 5 hours after template is added, and stored at 4°C in the mean time | |
| The quantity of the amplification product was not sufficient | Increase the number of PCR cycles to 35-40 to yield more amplification product | |
| Non-specific amplification product (s) | Annealing temperature was too low | Increase the annealing temperature |
| The number of PCR cycles was too high | Decrease the number of cycles to 30-35 | |
| Primer concentration was too high | Decrease primer concentration | |
| Template concentration was too high | Dilute template in purified H2O or TE buffer |
Tech Support
If you have any other enquiries, please leave a message.
* Indicates a Required Field







































